Histology (CIMA) dataset
This dataset became a part of the ANHIR - Automatic Non-rigid Histological Image Registration challenge which is part of ISBI 2019.
For an automatic evaluation (benchmark) of state-of-the-art or an automatic evaluation (benchmark) of state-of-the-art or your own methods, we recommend using BIRL: Benchmark on Image Registration methods with Landmark validations. This framework produces the final statistic as well as some mid-process visualisations.
The dataset consists of 2D histological microscopy tissue slices, stained with different stains, and landmarks denoting key-points in each slice. The task is image registration - align all slices in particular set of images (consecutive stain cuts) together, for instance to the initial image plane. The main challenges for these images are the following: very large image size, appearance differences, and lack of distinctive appearance objects. Our dataset contains 108 image pairs and manually placed landmarks for registration quality evaluation. A sample tissue of five stain cuts with highlighted landmarks is shown below.
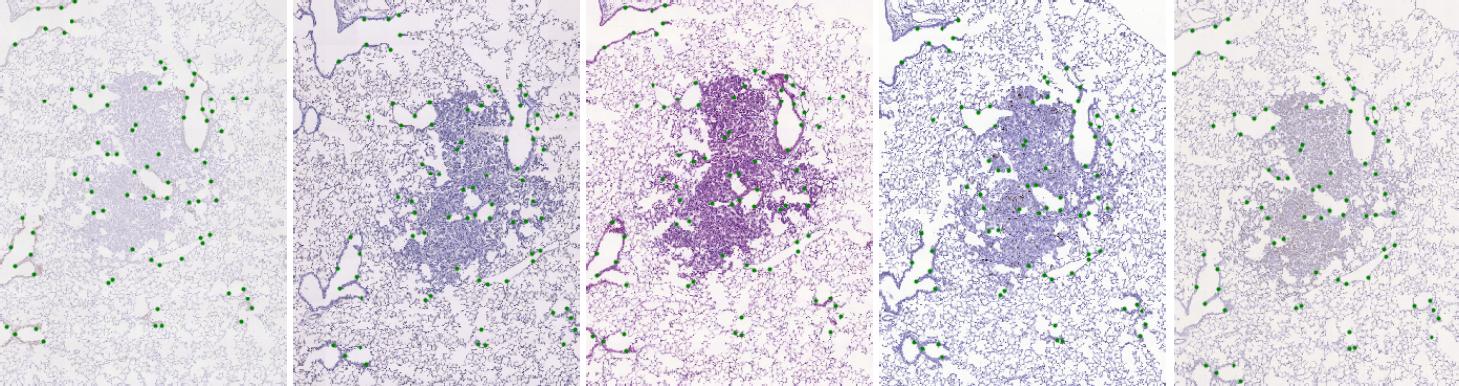
Dataset description
High-resolution (40x magnification) whole-slide images of a tissue (lesions, lung-lobes, mammary-gland) were acquired - the original size of our images varies have size up to 45k X 45k pixels. The acquired images are organized in 9 sets of consecutive sections where each slice was stained by a different dye and any two images within a set can be meaningfully registered.
The task is to register the images based on the tissue sections. The background can be ignored. We have marked significant structures in the tissue with landmarks which are spread approximately uniformly over the tissue. Moreover, the landmarks correspond to each other in the whole set of images acquired from the same tissue section, which allows us to validate the quality of the registration between any two images in the collection.
Hence, the dataset provides 108 image pairs to be registered together within the 9 sets.
The annotation of a single image pair took about 10 min and the whole set about 1 hour. We also provide downscaled versions of the images to 100%, 50%, 25%, 10% and 5%.
Consecutive tissue slices were stained with several different stains, see, see Stains and Molecular Markers.
The stains used are the following:
- Clara cell 10 protein (Cc10)
- Prosurfactant protein C (proSPC)
- Hematoxylin and Eosin (H&E)
- Antigen KI-67 (Ki67)
- Platelet endothelial cell adhesion molecule (PECAM-1, also known as CD31)
- Human epidermal growth factor receptor 2 (c-erbB-2/HER-2-neu)
- Estrogen receptor (ER)
- Progesterone receptor (PR)
Dataset summary
A brief summary of the dataset listing the particular sets with some statistical parameters.
| Dataset | Stains | Nb images | Nb landmarks | Avg. image size |
|---|---|---|---|---|
| Lesions 1 | Cc10, CD31, H&E, Ki67, proSPC | 5 | 78 | 16k |
| Lesions 2 | Cc10, CD31, H&E, Ki67, proSPC | 5 | 101 | 23k |
| Lesions 3 | Cc10, CD31, H&E, Ki67, proSPC | 5 | 80 | 16k |
| Lung lobes 1 | Cc10, CD31, H&E, Ki67, proSPC | 5 | 98 | 10k |
| Lung lobes 2 | Cc10, CD31, H&E, Ki67, proSPC | 5 | 107 | 10k |
| Lung lobes 3 | Cc10, CD31, H&E, Ki67, proSPC | 5 | 80 | 9k |
| Lung lobes 4 | Cc10, CD31, H&E, Ki67, proSPC | 5 | 86 | 9k |
| Mammary gland 1 | CNEU (=HER2), H&E, ER, PR | 5 | 82 | 22k |
| Mammary gland 2 | CNEU(=HER2), H&E, ER, PP | 8 | 76 | 20k |
| PAIRS | 9 (unique stains) | 108 | ~87 | - |
Pairing images
An example of the 10 registration pairs from a set of 5 differently stained images. Duplicates are omitted
| Cc10 | CD31 | H&E | Ki67 | proSPC | |
| Cc10 | - | 1 | 2 | 3 | 4 |
| CD31 | - | - | 5 | 6 | 7 |
| H&E | - | - | - | 8 | 9 |
| Ki67 | - | - | - | - | 10 |
| proSPC | - | - | - | - | - |
Landmarks
The landmarks are stored in CSV format which is very intuitive and can be used directly in ImageJ. The origin (0, 0) of the coordinate system is set to the image top left corner.
Further landmarks are available in a GitHub, which also provides the possibility to add new landmarks and share them among all dataset users. We highly appreciate any contributions increasing the overall landmark coverage.
Structure
The dataset is organized as follows: each set is stored in a separate folder, which contains a subfolder for each scale. In these, the images (at the particular scale) and the associated landmarks are stored, sharing the same file name. The images may be in PNG or JPG format, and the landmarks are in CSV.
DATASET |- [set_name1] | |- scale-5pc | | |- [image_name1].jpg | | |- [image_name1].csv | | |- [image_name2].jpg | | |- [image_name2].csv | | | ... | | |- [image_name].jpg | | '- [image_name].csv | |- scale-10pc | | ... | '- scale-100pc | |- [image_name1].png | |- [image_name1].csv | | ... | |- [image_name].png | '- [image_name].csv |- [set_name2] | ... '- [set_name]
License
If you use the images or the framework in your research work, we kindly ask you to cite our related contributions [1, 2, 3]. This work is made available under CC-BY-SA.Data acknowledgement
The images were provided by Prof. Arrate Munoz-Barrutia, Center for Applied Medical Research (CIMA), University of Navarra, Pamplona Spain [2]; and Prof. Ortiz de Solórzano, Center for Applied Medical Research (CIMA), University of Navarra, Pamplona Spain [3].Download
Please fill out the complete form, you will receive mail with a download link.
Preview
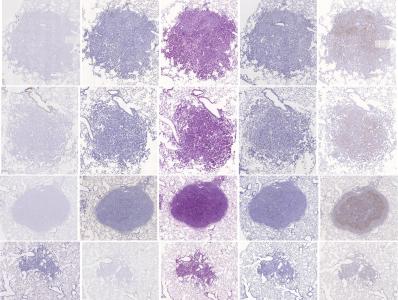
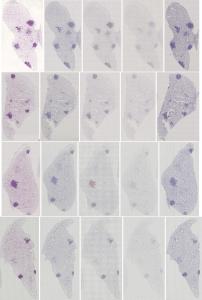
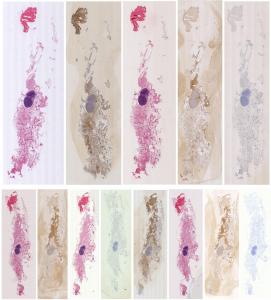
References
[1] J. Borovec, A. Munoz-Barrutia, and J. Kybic, “Benchmarking of Image Registration Methods for Differently Stained Histological Slides,” in IEEE International Conference on Image Processing (ICIP), 2018, pp. 3368–3372.[2] J. Borovec, J. Kybic, M. Bušta, C. Ortiz-de-Solorzano, and A. Munoz-Barrutia, “Registration of multiple stained histological sections,” in IEEE International Symposium on Biomedical Imaging (ISBI), 2013, pp. 1034–1037.
[3] R. Fernandez-Gonzalez et al., “System for combined three-dimensional morphological and molecular analysis of thick tissue specimens,” Microsc. Res. Tech., vol. 59, no. 6, pp. 522–530, 2002.



