Here is a list of my current and past research projects. It currently includes only some of the small projects my students were working on for their master, bachelor and other short term projects.
Presentation of our team is also on the faculty web page
Contents
- Geometric graph matching
- Microscopy image analysis
- Analysis of microscopy images of Langerhans islets
- Analysis of ultrasound images of the knee
- Detection of myeloma from low-dose CT images
- Automatic analysis of spatial gene expression patterns for Drosophila
- Registration of histology slices
- Elastic image registration
- Image registration uncertainty estimation
- High dimensional mutual information estimation
- Tool detection in 3D ultrasound
- Variational reconstruction
- Ultrasound elastography
- Detection of lung nodules from CT
- Parallel MRI reconstruction
- MEG/EEG reconstruction
- Respiratory motion estimation
- 3D medical image segmentation
- Automatic malaria diagnosis from microscopy images
- Opacity quantification in cardiac angiogram sequence
- Detection of impurities in oil in microscopy images
- Analysis of images from gel electrophoresis
- Colonoscopy image analysis
- Kalman filtering and speech enhancement
- Glowacki P., Pinheiro M. A., Mosinska A., Türetken E., Lebrecht D., Sznitman R., Holtmaat A., Kybic J. and Fua P.. “Reconstructing Evolving Tree Structures in Time Lapse Sequences by Enforcing Time-Consistency.” Trans. Pattern Analysis Machine Intelligence, vol. 40, no. 3, pp. 755-761, March 2018.
- Pinheiro M. A., Kybic J. and Fua P.. “Geometric Graph Matching Using Monte Carlo Tree Search.” Trans. Pattern Analysis Machine Intelligence, vol. 39, no. 11, pp. 2171-2185, 2017.
- Serradell Eduard, Amável Pinheiro Miguel, Sznitman Raphael, Kybic Jan, Moreno-Noguer Francesc and Fua Pascal. “Non-Rigid Graph Registration using Active Testing Search.” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 37, no. 3, pp. 625-638, March 2015.
- Habart David, Švihlík Jan, Schier Jan, Cahová Monika, Girman Peter, Zacharovová Klára, Berková Zuzana, Kříž Jan, Fabryová Eva, Kosinová Lucie, Papáčková Zuzana, Kybic Jan and Saudek František. “Automated Analysis of Microscopic Images of Isolated Pancreatic Islets.” Cell Transplantation, no. 12, pp. 2145-2156, December 2016.
- Svihlik J., Kybic J. and Habart D.. “Color normalization for robust evaluation of microscopy images.” Optics and Photonics 2015: Applications of Digital Image Processing XXXVIII, vol. 9599, pp. 1-6, August 2015.
- Švihlík J., Kybic J., Habart D., Berková Z., Girman P., Kříž J. and Zacharovová K.. “Classification of microscopy images of Langerhans islets.” Medical Imaging 2014: Image Processing, vol. 9034, pp. 1-8, February 2014.
- Martínez F., Kybic J., Lambert L. and Mecková Z.. “Fully-automated classification of bone marrow infiltration in low-dose CT of patients with multiple myeloma based on probabilistic density model and supervised learning.” Computers in Biology and Medicine, vol. 71, pp. 57-66, April 2016.
- Francisco Martínez-Martínez, Jan Kybic and Lukas Lambert. “Automatic detection of bone marrow infiltration by multiple myeloma detection in low-dose CT.” International Conference in Image Processing (ICIP), vol. , no. , pp. 4813-4817, September 2015.
- Borovec J., Kybic J. and Sugimoto A.. “Region growing using superpixels with learned shape prior.” Journal of Electronic Imaging, 2017.
- Borovec J., Švihlík J., Kybic J. and Habart D.. “Supervised and unsupervised segmentation using superpixels, model estimation, and Graph Cut.” Journal of Electronic Imaging, 2017.
- Borovec J., Kybic J. and Nava R.. “Detection and localization of Drosophila egg chambers in microscopy images.” MLMI: Machine learning in medical imaging (MICCAI workshop), pp. 19-26, September 2017.(Accepted)
- Nava Rodrigo and Kybic Jan. “Supertexton-based segmentation in early Drosophila oogenesis.” International Conference in Image Processing (ICIP), pp. 2656-2659, 2015.
- Jan Kybic and Jiri Borovec. “Automatic simultaneous segmentation and fast registration of histological images.” International Symposium on Biomedical Imaging (ISBI), pp. 774-777, April-May 2014.
- Jan Kybic. “Registration of segmented histological images using thin plate splines and belief propagation.” SPIE Medical Imaging, vol. 9034, pp. 90343E, February 2014.(Accepted)
- Ledesma-Carbayo María Jesús, Jan Kybic, Manuel Desco, Andrés Santos, Michael Sühling, Patrick Hunziker and Michael Unser. “Spatio-Temporal Non-Rigid Registration for Ultrasound Cardiac Motion Estimation.” IEEE Transactions on Medical Imaging, vol. 24, no. 9, pp. 1113-1126, September 2005.
- Jan Kybic and Michael Unser. “Fast Parametric Elastic Image Registration.” IEEE Transactions on Image Processing, vol. 12, no. 11, pp. 1427-1442, November 2003.
- Jan Kybic, Philippe Thévenaz, Arto Nirkko and Michael Unser. “Unwarping of Unidirectionally Distorted EPI Images.” IEEE Transactions on Medical Imaging, vol. 19, no. 2, pp. 80-93, February 2000.
- Jan Kybic. “Biomedical Image Processing by Elastic Warping.” July 2001.
- Kybic Jan and Nieuwenhuis Claudia. “Bootstrap Optical Flow Confidence and Uncertainty Measure.” Computer Vision and Image Understanding, vol. 115, no. 10, pp. 1449-1462, June 2011.
- Jan Kybic. “Bootstrap Resampling for Image Registration Uncertainty Estimation without Ground Truth.” IEEE Trans. Image Processing, vol. 19, no. 1, pp. 64-73, January 2010.
- Kybic Jan and Vnučko Ivan. “Approximate All Nearest Neighbor Search for High Dimensional Entropy Estimation for Image Registration.” Signal Processing, vol. 92, no. 5, pp. 1302-1316, 2012.
- García-Arteaga Juan David, Kybic Jan and Li Wenjing. “Automatic Colposcopy Video Tissue Classification Using Higher Order Entropy Based Registration.” Computers in Biology and Medicine, vol. 41, no. 10, pp. 960-970, October 2011.
- Kybic Jan. “High-Dimensional Entropy Estimation for Finite Accuracy Data: R-NN Entropy Estimator.” IPMI2007: Information Processing in Medical Imaging, 20th International Conference, pp. 569-580, July 2007.
- Jan Kybic. “Incremental updating of nearest neighbor-based high-dimensional entropy estimation.” ICASSP2006, pp. III-804, May 2006.
- Jan Kybic. “High-Dimensional Mutual Information Estimation For Image Registration.” ICIP‘04: Proceedings of the 2004 IEEE International Conference on Image Processing, October 2004.
- García-Arteaga Juan David. “Multichannel Image Information Similarity Measures: Applications to Colposcopy Image Registration.” pp. 120, May 2013.
- Marián Uherčík, Jan Kybic, Hervé Liebgott and Christian Cachard. “Model Fitting Using RANSAC for Surgical Tool Localization in 3-D Ultrasound Images.” IEEE Transactions on Biomedical Engineering, vol. 57, no. 8, pp. 1907-1916, Aug. 2010.
- Martin Barva, Marián Uherčík, Jean-Martial Mari, Jan Kybic, Jean-René Duhamel, Hervé Liebgott, Václav Hlaváč and Christian Cachard. “Parallel Integral Projection Transform for Straight Electrode Localization in 3-D Ultrasound Images.” IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control (UFFC), vol. 55, no. 7, pp. 1559-1569, July 2008.
- Marián Uherčík, Jan Kybic, Christian Cachard and Hervé Liebgott. “Line filtering for detection of microtools in 3D ultrasound data.” Ultrasonics Symposium (IUS), 2009 IEEE International, pp. 594-597, September 2009.
- Marián Uherčík. “Surgical Tools Localization in 3D Ultrasound Images.” pp. 135, April 2011.
- Barva Martin. “Localization of Surgical Instruments in 3D Ultrasound Images.” pp. 153, June 2007.
- Jan Kybic, Thierry Blu and Michael Unser. “Generalized sampling: A variational approach. Part II - Applications.” IEEE Transactions on Signal Processing, vol. 50, no. 8, pp. 1977-1985, August 2002.
- Jan Kybic, Thierry Blu and Michael Unser. “Generalized sampling: A variational approach. Part I - Theory.” IEEE Transactions on Signal Processing, vol. 50, no. 8, pp. 1965-1976, August 2002.
- Jan Kybic. “Biomedical Image Processing by Elastic Warping.” July 2001.
- Jan Kybic and Daniel Smutek. “Computational Elastography from Standard Ultrasound Image Sequences by Global Trust Region Optimization.” Proceedings of IPMI, Lecture Notes in Computer Science, no. 3565, pp. 299-310, July 2005.
- Jan Kybic and Daniel Smutek. “Estimating Elastic Properties of Tissues from Standard 2D Ultrasound Images.” SPIE Medical Imaging 2005: Ultrasonic Imaging and Signal Processing, vol. 6, no. 5750, pp. 184-195, February 2005.
- Neuwirth J., Polovinčák M., Kybic J., Tůma S., Čumlivská E., Suchánek V., Adla T., Hlaváč V. and Dolejší M.. “Solitární a mnohočetné plicní uzly - analýza morfologických vlastností na průkaz jejich etiologie, pravidla sledování malých uzlů, jejich detekce pomocí semiautomatické analýzy CT obrazu - vlastní zkušenosti a přehled literatury.” Česká radiologie, vol. 60, no. 5, pp. 311-320, March 2006.
- Dolejší Martin, Kybic Jan, Polovinčák Michal and Tůma Stanislav. “Reducing false positive responses in lung nodule detector system by Asymmetric AdaBoost.” Proceedings of 2008 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, pp. 656-659, May 2008.
- Dolejší Martin and Kybic Jan. “Automatic two-step detection of pulmonary nodules.” Proceedings of SPIE, vol. 6514, pp. 3j-1-3j-12, February 2007.
- Petr Jan, Kybic Jan, Bock Michael, Müller Sven and Hlaváč Václav. “Parallel Image Reconstruction Using B-Spline Approximation (PROBER).” Magnetic Resonance in Medicine, vol. 58, no. 9, pp. 582-591, September 2007.
- Petr Jan. “Parallel Magnetic Resonance Imaging Reconstruction.” pp. 129, October 2007.
- Clerc Maureen and Kybic Jan. “Cortical mapping by Laplace-Cauchy transmission using a boundary element method..” Journal on Inverse Problems, vol. 23, pp. 2589-2601, November 2007.
- Jan Kybic, Maureen Clerc, Olivier Faugeras, Renaud Keriven and Théo Papadopoulo. “Generalized Head Models for MEG/EEG: BEM beyond Nested Volumes.” Physics in Medicine and Biology, vol. 51, no. 5, pp. 1333-1346, February 2006.
- Jan Kybic, Maureen Clerc, Olivier Faugeras, Renaud Keriven and Théo Papadopoulo. “Fast Multipole Acceleration of the MEG/EEG Boundary Element Method.” Physics in Medicine and Biology, vol. 50, no. 19, pp. 4695-4710, October 2005.
- Kybic Jan, Clerc Maureen, Abboud Touffic, Faugeras Olivier, Keriven Renaud and Papadopoulo Théo. “A common formalism for the integral formulations of the forward EEG problem.” IEEE Transactions on Medical Imaging, vol. 24, no. 1, pp. 12-28, January 2005.
- Jef Vandemeulebroucke, Jan Kybic, Patrick Clarysse and David Sarrut. “Respiratory Motion Estimation from Cone-Beam Projections Using a Prior Model.” Medical Image Computing and Computer-Assisted Intervention - MICCAI 2009, vol. 5762, pp. 365-372, September 2009.
- Jef Vandemeulebroucke, Patrick Clarysse, Jan Kybic and David Sarrut. “Estimating Respiratory Motion from Cone-Beam Projections.” Proceedings of the First International Workshop on Pulmonary Image Analysis, pp. 83-92, September 2008.
- Jef Vandemeulebroucke. “Lung Motion Modelling and Estimation for Image Guided Radiation Therapy.” pp. 148, July 2010.
- Jan Kybic and Jakub Krátký. “Discrete curvature calculation for fast level set segmentation.” ICIP: International Conference on Image Processing, pp. 4404, November 2009.
- Krátký Jakub and Kybic Jan. “Three-dimensional segmentation of bones from CT and MRI using fast level sets.” SPIE MI 2008: Proceedings of the SPIE Medical Imaging 2008 Conference, vol. 6914, pp. 10, February 2008.
- Vít Špringl. “Automatic Malaria Diagnosis through Microscopy Imaging.” pp. 127, February 2009.
- Kazmar T. and Kybic J.. “Opacity Quantification In Cardiac Angiogram Sequences.” BIOSIGNAL: Analysis of Biomedical Signals and Images, pp. 66, June 2008.
- Jan Kybic. “Kalman filtering and speech enhancement.” 1998.
Research projects
Geometric graph matching
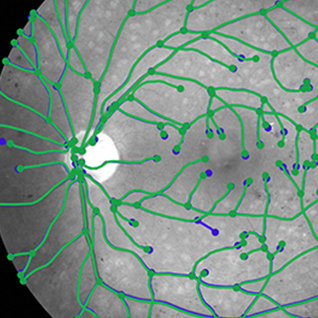 |
We solve the problem of efficient matching method for generalized geometric graphs. Such graphs consist of vertices in space connected by curves and can represent many real world structures such as road networks in remote sensing, or vessel networks in medical imaging. Graph matching can be used for very fast and possibly multimodal registration of images of these structures. Our method is very efficient — it can match graphs with thousands of nodes, which is an order of magnitude better than the best competing method. Participants: Miguel Amável Pinheiro. Selected publications: |
Microscopy image analysis
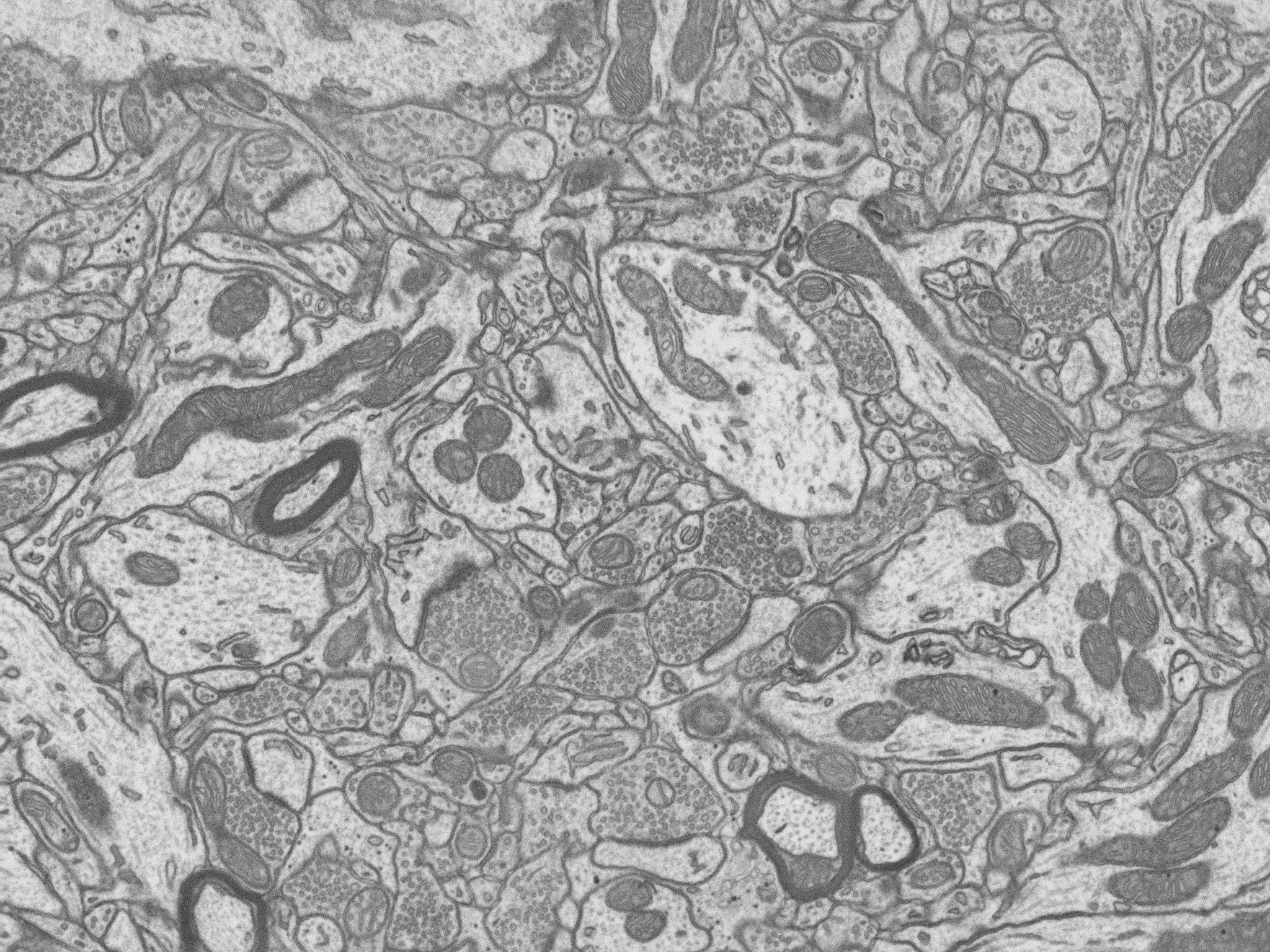 |
We are developing algorithms for tracking of neuronal fibers (processes) in electron microscopy (especially 3D, FIB) and light microscopy (bright field and confocal) images and extracting the neuronal trees. Our main focus is alignment of electron microscopy and optical microscopy volumes. Participants: Miguel Amável Pinheiro, Thomas Dietenbeck. |
Analysis of microscopy images of Langerhans islets
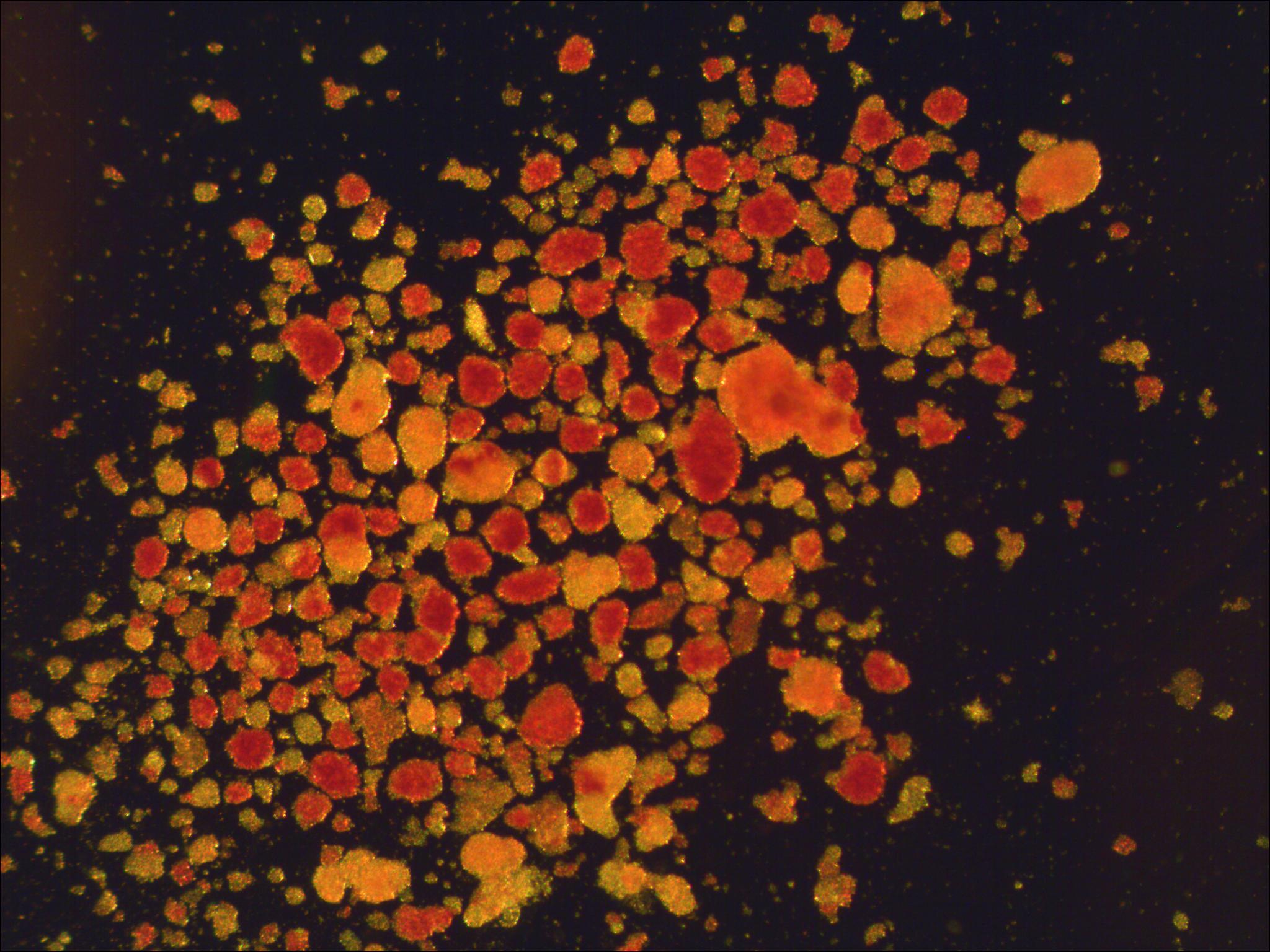 |
In collaboration with IKEM, we are developing automatic techniques to analyze microscopic images of Langerhans islets to determine their suitability for transplantation. Participants: Jan Švihlík. Selected publications: |
Analysis of ultrasound images of the knee
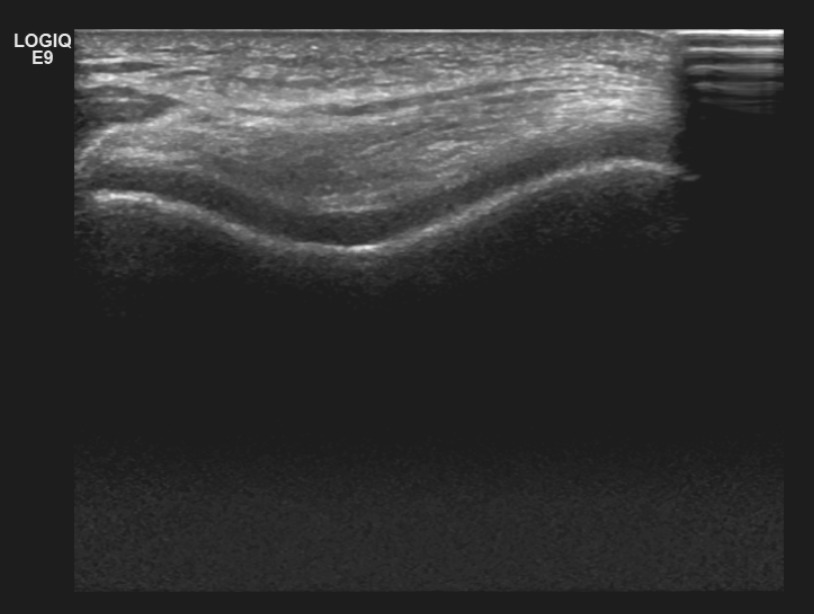 |
We work on the analysis of ultrasound images of the knee to diagnose osteoarthritis. |
Detection of myeloma from low-dose CT images
 |
We attempt to detect multiple myeloma from low-dose whole-body CT images. We focus on the long bones, especially femur. So far, there is no automatic way to perform this analysis. This is a collaboration with the Faculty of Medicine, Charles University. Participants: Francisco Martínez, Jan Hering. Selected publications: |
Automatic analysis of spatial gene expression patterns for Drosophila
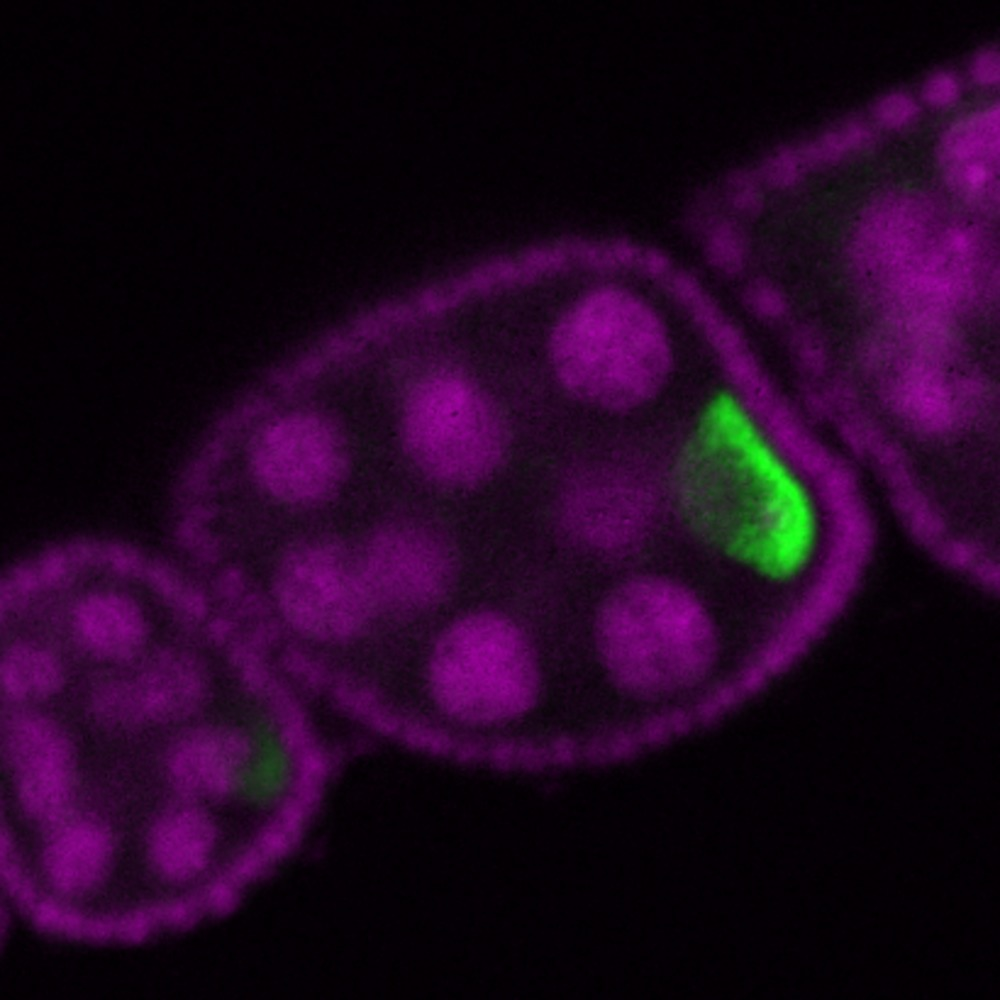 |
The aim of this project is to develop a set of algorithmic tools for processing large sets of microscopy images for systematic survey of spatio-temporal gene expression patterns of Drosophila ovaries and larval imaginal disc. The key idea is to automatically group genes with similar spatio-temporal expressions in order to decipher the gene regulatory networks underlying the patterning of the tissues, with applications for modeling human disease conditions. This is a collaboration with the Max Planck institute in Dresden. Segmentation software pyImSegm released. Participants: Rodrigo Uriel Nava, Jiří Borovec. Selected publications: |
Registration of histology slices
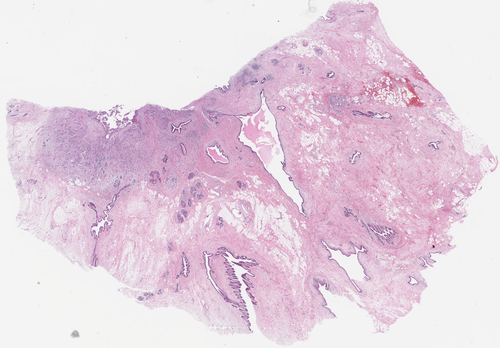 |
We are developing algorithms for registration of histology slices with different stainings. The challenge is the large size of the images and the variability of their appearances. Our idea is to combine segmentation and registration. Participants: Jiří Borovec, Martin Dolejší. Selected publications: |
Elastic image registration
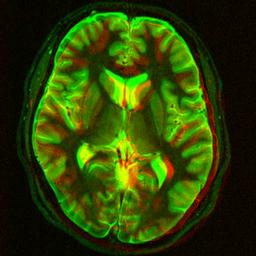 |
Image registration is the task of finding a geometrical transformation between two images of the same or similar object. In biomedical imaging, registration is often used to detect and quantify movement or changes, and to align images taken at different times or using different imaging devices. In my PhD work I have developed an elastic registration algorithm using B-splines which I have applied to several practical problems. My original implementation in Matlab, and Python+C is available, there is now also an improved version. There is also a closely related code (not written by me) in C++ which is now part of ITK and an ImageJ and Fiji Java plugin. Participants: Ignacio Arganda-Carreras. Selected publications: |
Image registration uncertainty estimation
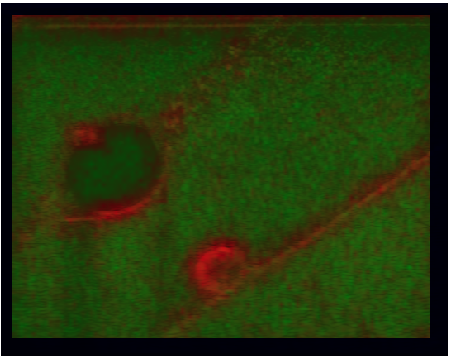 |
Standard image registration algorithms give us a single, deterministic answer. Here we are addressing the problem of estimating its accuracy, how much the results can be trusted. Our method is based on bootstrap resampling and needs nothing more than the two input images and the registration algorithm. Selected publications: |
High dimensional mutual information estimation
 |
When two image from different modalities are to be registered, the criterion of choice is mutual information, which can adapt itself to any dependence between the intensities in the two images. Sometimes it might be useful to use not only scalar features (such as image intensities) but vector features, such as colour, neighborhood information, or texture information. However, this is difficult to do with standard mutual information implementations based on histograms, which do not work well in higher dimensions. We are therefore studying alternative estimators based on Kozachenko-Leonenko nearest neighbor estimators. Since finding all the nearest neighbors is the computational bottleneck, we have a practical algorithm using kD-trees, best bin first strategy and incremental updating. Participants: Juan David García-Arteaga, Jiří Svoboda, Sudershan Sundararajan. Selected publications: |
Tool detection in 3D ultrasound
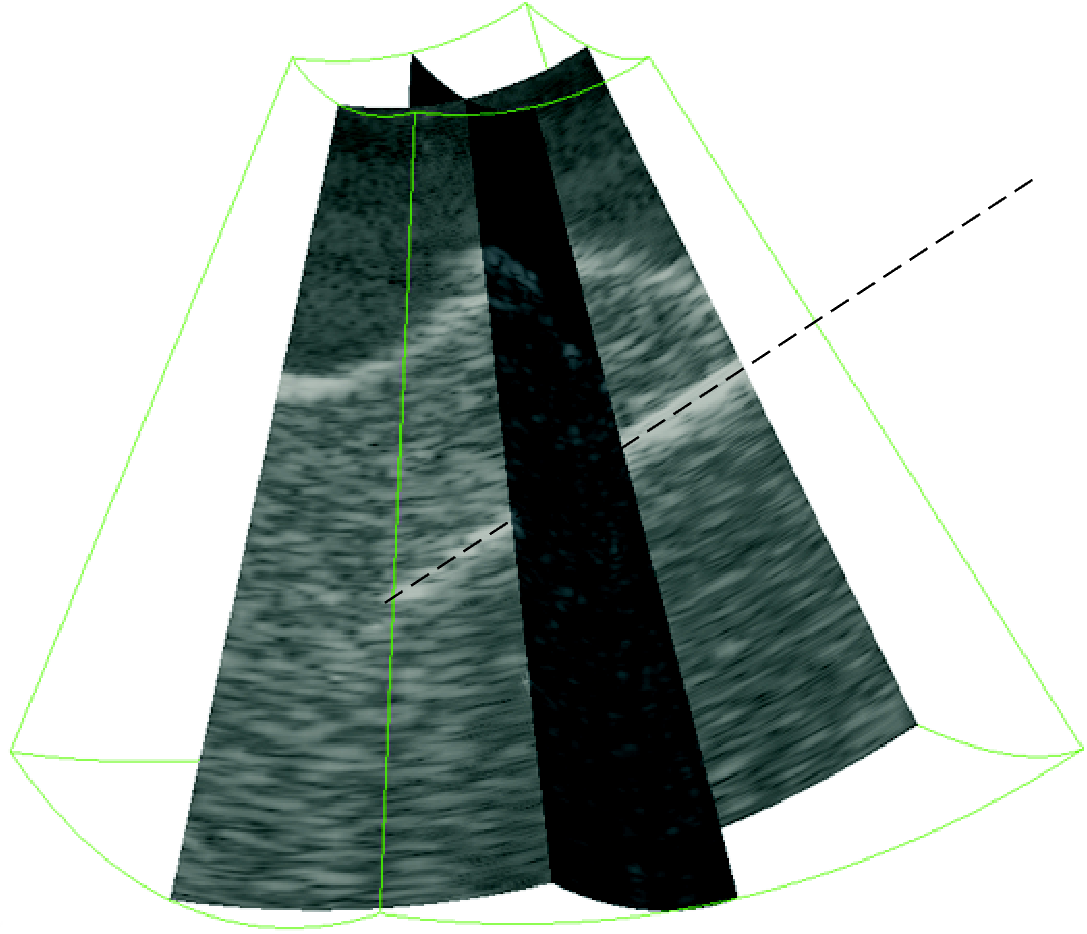 |
We solve the task of fast and robust detection and localization of surgical tools such as needles or electrodes from 3D ultrasound images for visualization and automatic guidance purposes. We have proposed a localization method based on Parallel Integral Projection and another one based on RANSAC (Random Sampling Consensus). There is also a demonstration application. This project was done in collaboration with CREATIS, INSA Lyon, France. Participants: Martin Barva, Marián Uherčík. Selected publications: |
Variational reconstruction
 |
We consider the problem of reconstruction a multidimensional and multivariate function from a finite set of linear measures, so that the reconstruction is optimal in the sense some quadratic regularization criterion. This is also called a generalized sampling and generalized interpolation problem. A simple is to interpolate a function given its values and derivatives, so that the norm of its second derivative is minimal. Another example is to recover a 2D density function given its integral projections (i.e. the Radon transform) so that the reconstruction is as smooth as possible, e.g. in the sense of a second-order Duchon’s seminorm (almost a Laplacian). We show that imposing generally desirable properties on the reconstruction largely limits the choice of the regularization criterion and that the solution can be obtained as a linear combination of generating functions, which can be found analytically. Selected publications: |
Ultrasound elastography
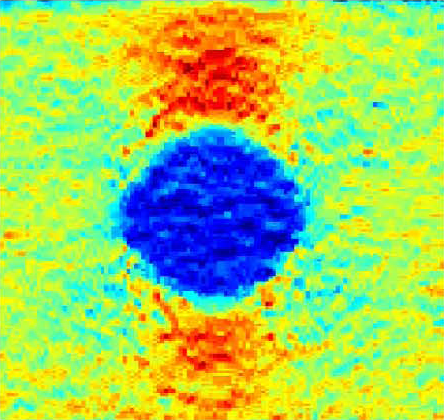 |
Ultrasound elastography attempts to reconstruct pointwise mechanical properties of the tissue from an ultrasound sequence. To put it simply, if a tissue deforms a lot, it is probably soft and vice versa. This can be useful for medical diagnostics of cancer in soft tissue close to the surface, for example in the breast, thyroid gland or liver, since many tumors are harder than the surrounding tissue. Analyzing the stiffness of vessels, such as the carotid artery, may be useful for the diagnostics of atherosclerosis. We have proposed several reconstruction algorithms for standard ultrasound image sequences and we also have the opportunity to work with raw RF (radio-frequency) signal which allows to recover the motion better. We address the computational problem of reconstructing the elasticity properties from the images. We have our own ultrasound scanner with RF output. This project is in collaboration with CREATIS, INSA Lyon, France and the First Faculty of Medicine, Charles University, Prague. Selected publications: |
Detection of lung nodules from CT
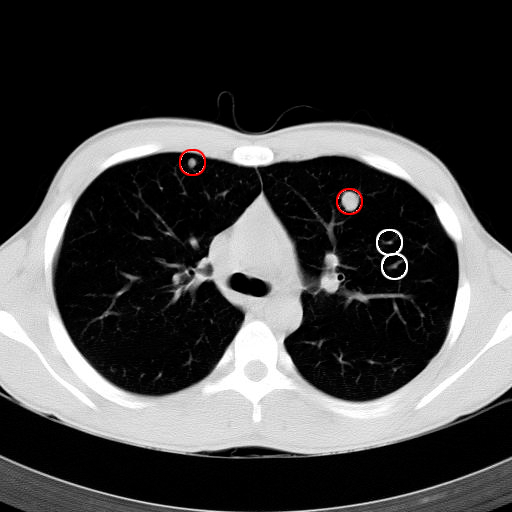 |
Pulmonary nodules are small lesions in the lungs, which may indicate a lung cancer. They are best detected from CT (computed tomography) 3D scans. Their early detection significantly improves patients’ survival rate. We are working on an automatic nodule detection system to identify suspected nodules. It should serve as a tool for a radiologist, in order to minimize false negatives (overlooked nodules). We have developed a classification-based algorithm and we have also assembled a large annotated image database. This work is done in collaboration with the Motol Hospital in Prague.Participants: Martin Dolejší. Selected publications: |
Parallel MRI reconstruction
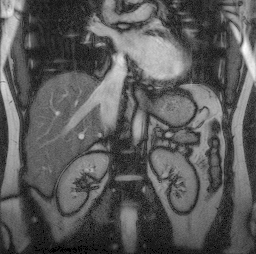 |
Parallel MRI (magnetic resonance imaging) uses several acquisition coils instead of just one. This allows to accelerate the acquisition which is important for imaging dynamic sequences, reduce motion artifacts, reduce patient discomfort and increase the throughput of the scanner. However, the reconstruction technique is more complicated than in the standard case, as the sub-sampling leads to aliasing artifacts which must be compensated using differences in spatial sensitivity of the coils. We have developed a new parallel MRI reconstruction method called PROBER using B-splines to take advantage of the spatial smoothness of the sensitivity fields. Our method provides better reconstruction quality than standard SENSE and GRAPPA reconstruction methods. This work was done in collaboration with DKFZ, Heidelberg, Germany. Participants: Jan Petr. Selected publications: |
MEG/EEG reconstruction
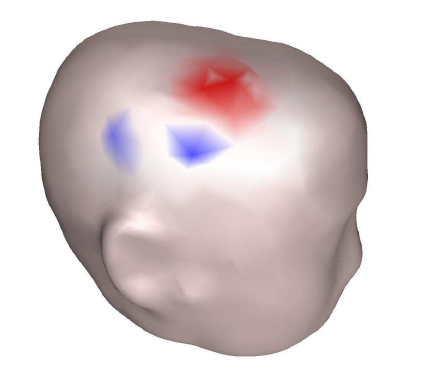 |
Magnetoencephalography (MEG) and electroencephalography (EEG) measure small magnetic and electric fields on the scalp caused by neuronal activity. We address the computational issues of reconstructing the spatio-temporal neuronal activity from these measurements. We have worked mainly on the numerical aspects of the forward modeling using finite element method (FEM) and boundary element method (BEM), adapting a little known symmetric variant of the BEM to our problem and generalizing it to complicated topologies, as well as accelerating the computation using a fast multipole method (FMM). Another topic of interest is the solvability and uniqueness of the inverse problem. This work was done in collaboration with the Odyssée group at INRIA, Sophia-Antipolis, France. Selected publications: |
Respiratory motion estimation
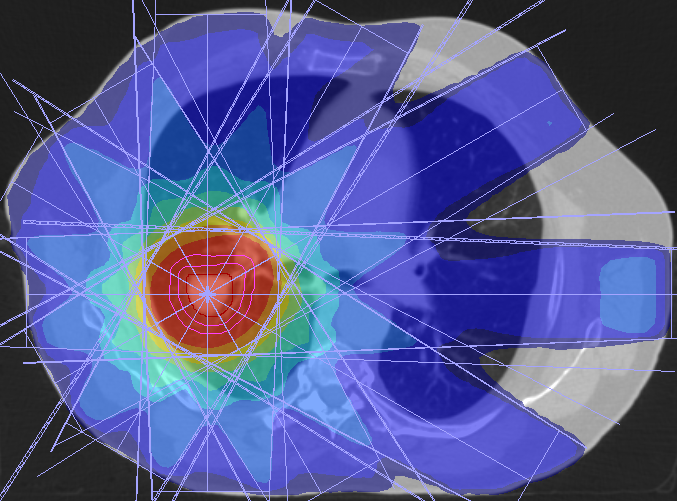 |
Radiation therapy consists of exposing tumor cells to ionizing radiation. Breathing motion during irradiation should be taken into account during planning and ideally also the treatment itself. It is therefore important to model the breathing motion well. Our model is patient-specific and is based on preoperatively acquired 4D CT sequences, elastic image registration and B-spline information. During treatment, the model is adapted using the available measurements. Participants: Jef Vandemeulebroucke. Selected publications: |
3D medical image segmentation
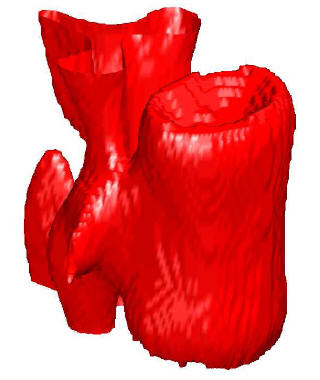 |
Segmentation of medical images is one of the most useful and most often used operations. Our main application is segmentation of bones and cartilages for biomechanical modeling. We have experimented with GraphCut methods but we mostly focus on fast level set methods which use a discrete formulation instead of solving PDE and are therefore very fast. More speedup can be achieved by our discrete method of curvature estimation with online updating. This project is in collaboration with Faculty of Mechanical Engineering of the Czech Technical University. Participants: Jakub Krátký. Selected publications: |
Automatic malaria diagnosis from microscopy images
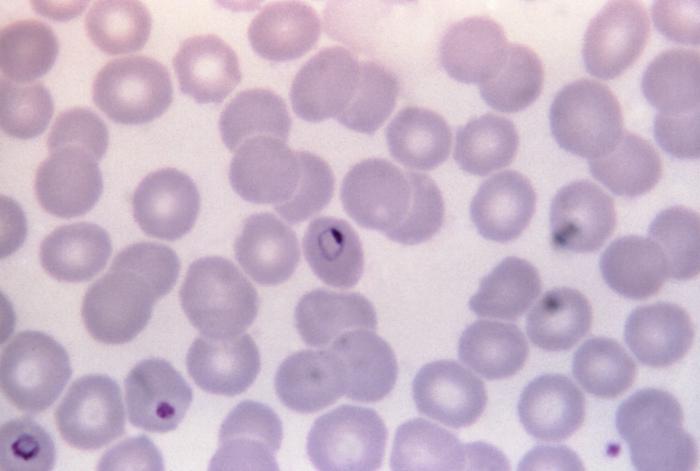 |
The project (started by Juan David García) deals with processing microscopy images for automatic malaria diagnostics. First individual cells have to be segmented and then the presence of parasites is detected. Participants: Juan David García-Arteaga, Vít Špringl. Selected publications: |
Opacity quantification in cardiac angiogram sequence
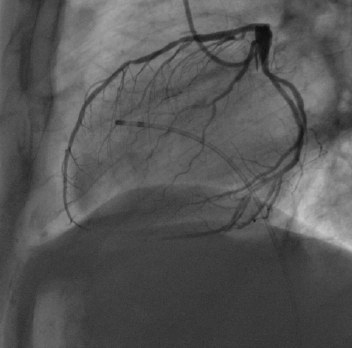 |
The goal of this project is to automatically analyze cardiac angiogram X-ray sequences to evaluate the perfusion for diagnosis of microvascular dysfunction. The method involves image registration for movement compensation and compartmental modelling for perfusion analysis. Participants: Tomáš Kazmar. Selected publications: |
Detection of impurities in oil in microscopy images
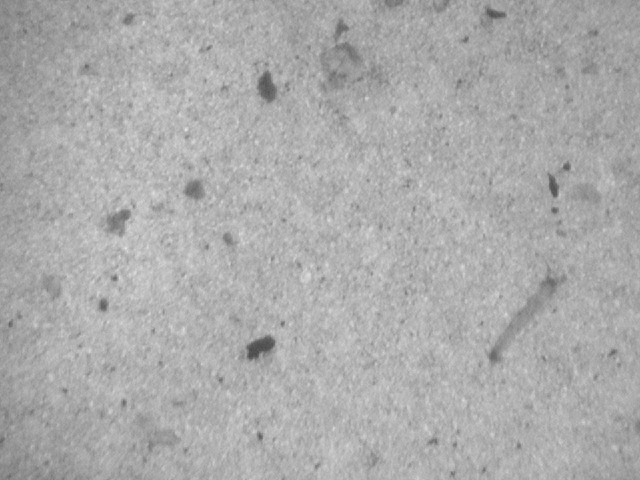 |
The idea is to analyze microscopy images of oil samples, detected possible imputirities and thus quantify the quality of the oil. This was a master thesis of Ivo Čermák, I was a consultant. Participants: Ivo Čermák. |
Analysis of images from gel electrophoresis
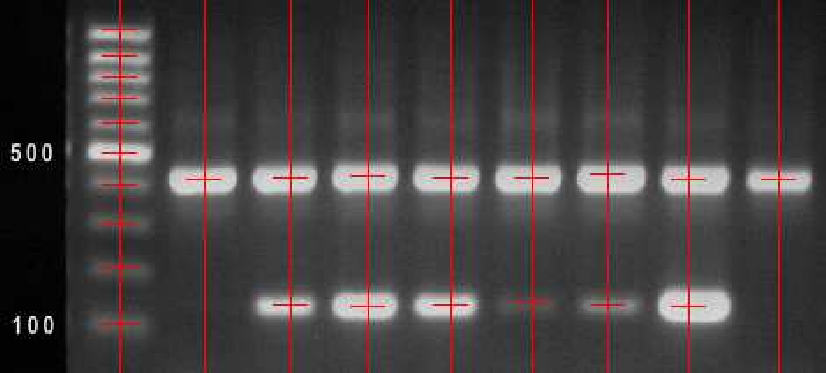 |
The task is to improve the image quality of gel electrophoresis images by noise filtering and to correct geometric distortion caused by field inhomogeneity. The next step is marker (spot) localization for automatically or semiautomatically evaluation of the electrophoresis results. Participants: Jiří Kaňka, Barbora Vrbová. |
Colonoscopy image analysis
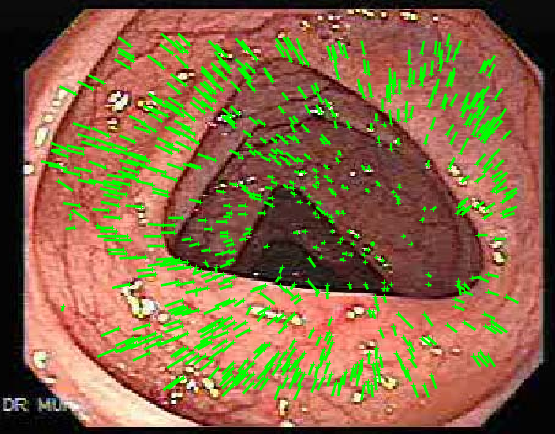 |
We attempted to reconstruct a 3D surface of the colon from a monocular colonoscopic video with uncalibrated camera for further modeling and to help diagnostics, such as polyp detection. Participants: Helena Jansová. |
Kalman filtering and speech enhancement
 |
In my master thesis (a long time ago) I have worked on the problem of suppressing car noise from speech signals for hands-free phone operations. I have used Kalman filtering and a speech detector to estimate the spectrum of the speech and the noise. Selected publications: |

